Epigenetic modifications during exercise
Фотографии:
ˑ:
PhD, Associate Professor N.D. Golberg1
PhD, Associate Professor I.V. Astratenkova2
Dr.Med. I.I. Akhmetov1
Dr.Biol., Professor V.A.Rogozkin1
1St. Petersburg Research Institute of Physical Culture, St. Petersburg
2St. Petersburg State University, St. Petersburg
Keywords: epigenetics, DNA methylation, modification of histones, microRNA, exercise.
The study of the molecular mechanisms of gene expression in skeletal muscles has been one of significant issues of molecular genetics, biochemistry and human physiology for many years. Until recently, you could find a dominant statement in genetics that the DNA is the only carrier of hereditary information. Mapping on the human genome and genome of other organisms revealed not only the DNA organization principles, but also showed the regulation complexity of the structural and metabolic genes expression. It turned out that external influences such as nutrition, stress, physical activity and other external stimuli change genes expression and control it through a complex set of regulatory mechanisms.
It became necessary to find more accurate concept definitions, and now besides the ‘genome’ – the term ‘epigenome’ has become a commonly used term which includes assembly of elements regulating genes expression. These elements do not confine to the primary DNA structure, but can be inherited. Epigenetic modifications include DNA and histones methylation, modifications of histones and regulatory proteins in the reactions of acetylation, phosphorylation, ubiquitylation, sumoylation, acetylglucosoamination and proteolytic digestion. Cascades of reactions associated with epigenetic post-translational modification of proteins are a universal mechanism for controlling intracellular metabolism, which provides differentiation, transformation, hypertrophy and atrophy of muscle cells.
Objective of the article was to show the development and current state of molecular genetics of human physical activity.
At the turn of the 21st century, articles published in this journal showed the presence of genetic markers in humans that determine physical performance [4,5]. Mapping on the human genome has opened wide possibilities for application of the genetic methods in various spheres of human activity including sport. Intensive study of gene polymorphisms responsible for predisposition to the athletic performance in different sports which were conducted in Saint Petersburg Research Institute of Physical Culture served as the basis for the formation of sports genetics in our country. The results of research were presented in several PhD theses and summarized in the doctoral dissertation of Akhmetov I.I. To popularize the new direction of science Akhmetov I.I. wrote a book "Molecular genetics of sports" in 2009 [3]. In this book the author provided a detailed description of development stages of sports molecular genetics in our country and abroad. Similar laboratories in Europe, USA, South America, China, Korea, Japan, South Africa and other countries were mentioned in his book.
Over the years, this scientific area has been developed and gradually shaped. Molecular sports genetics studies molecular mechanisms and patterns of human physical activity inheritance. The main direction of research is connected with the determination of the most common DNA polymorphisms associated with performance of various human physical and mental qualities during daily physical activity and professional athletic activities.
Currently, the search for genetic markers associated with a predisposition to various sports keeps on at the laboratories in different countries. The results of these studies have gone beyond the laboratories. The service companies have appeared offering genetic testing for children and adolescents in order to determine their genetic predisposition to various sports. The widespread commercialization of genetic services for the population raised concerns at the International Federation of Sports Medicine which made an official statement. In May of 2015 leading sports geneticists held a symposium and discussed the state and perspectives for the development of human genetics of physical activity. They prepared and published a collective project “Athlome” (Athlome Project) which defined four areas for further research [10]. The executive committee of “Athlome” includes 16 leading scientists including representatives of the Russian school of sports genetics.
Numerous genetic studies conducted in different countries have shown that the metabolism regulation in the human organism is represented not only by genes expression at the DNA level, but includes a more complex mechanism involving epigenetic factors. In addition, it became clear that besides the structural features of the DNA (polymorphisms, mutations), human predisposition to physical exercises is also affected by epigenetic modifications. These modifications are reversible processes by their nature and can have positive or negative influence on physical performance.
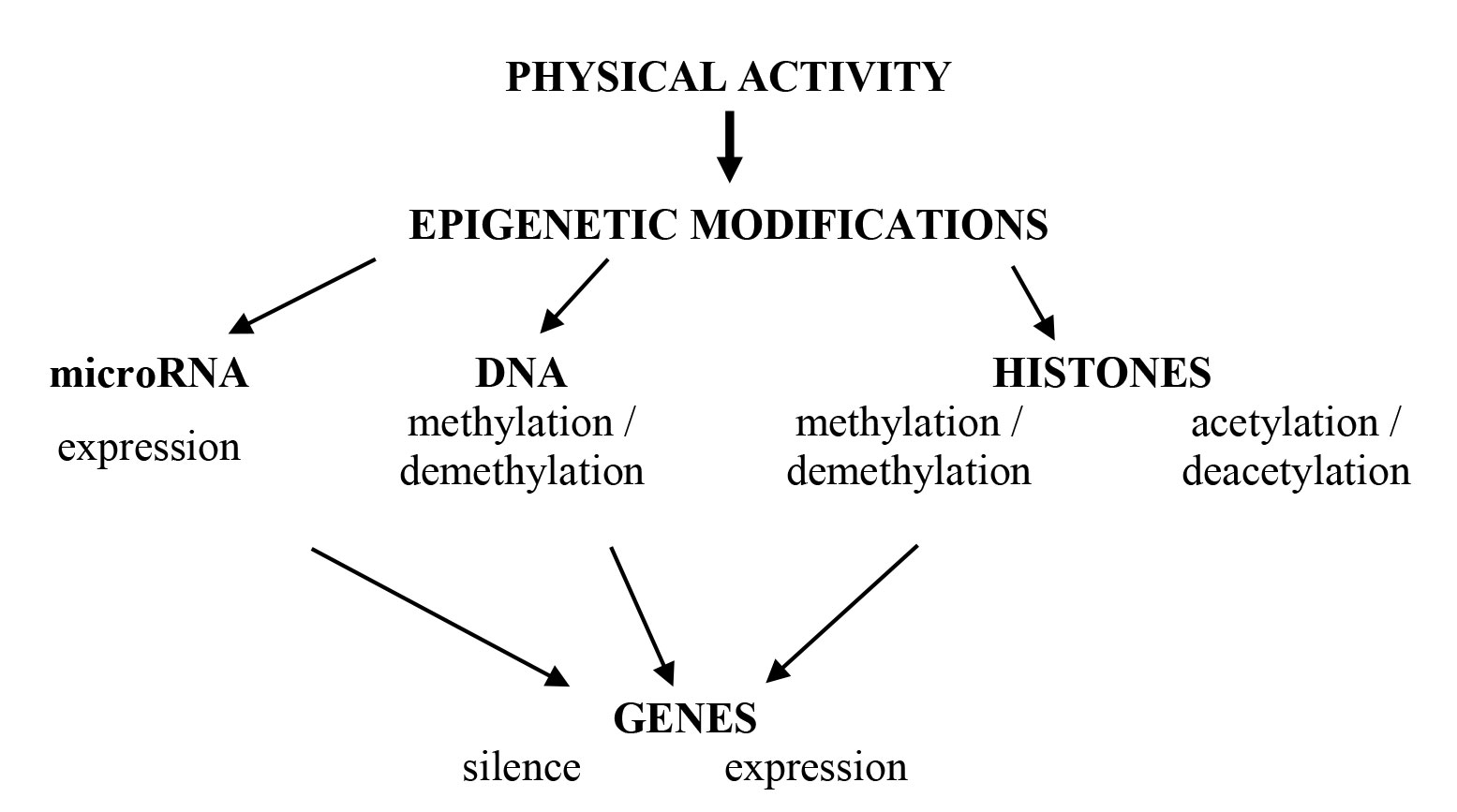
Let us briefly consider recent achievements in human epigenetics associated with physical performance. Participants of epigenetic regulation are shown on Figure 1.
Muscle cells have congenital and individual epigenetic features called an epigenome which defines two common epigenetic events: DNA methylation and histone modification. These processes control DNA conformation in two different states: ‘open’ with transcriptional active euchromatin and ‘closed’ with transcriptional inactive heterochromatin. In such a way, DNA participation in transcription, repair and replication is regulated.
Methylation is a temporary chemical modification of the nucleotide sequence without alteration of the DNA coding ability. In this case reversible methylation is considered as an epimutation unlike a mutation caused by nucleotide substitutions, lack of a gene site or, finally, insertion of nucleotides. The process of methylation and demethylation of the DNA in skeletal muscles is catalyzed by DNA methyltransferases (DNMT) and demethylases, which are called ten-eleven translocase enzymes (TET). In human skeletal muscles, DNA methylation process involves the transfer of the methyl group (-CH3) to the C5 nucleotide position of the cytosine, which catalyzes a DNMT family. DNMT family consists of three enzymes that differ in expression intensity during physical activity. Methylation occurs in CpG-islands, resulting in the formation of methyl CpG-sequences on both DNA chains. Among seven possible modifications of histones, only methylation and acetylation of histones in the skeletal muscles are considered in the article, since they are the ones that most intensively change during exercising.
Methylation of histones is more diverse and leads to different changes in the intensity of gene expression. Methylation of lysine 4 in histone H3 (H3K4me3) causes an increase in gene expression. Methylation of lysine at position 9 and 27 of histone H3 (H3K9me2 / 3 and H3K27me3) and lysine 20 in histone H4 (H4K20me3) inhibits gene expression. It is assumed that monomethylation of histones leads to activation of transcription processes, whereas di- and trimethylation of histones, on the contrary, is accompanied by inhibition of transcription. These processes are regulated in reactions of interaction between histone modification and DNA methylation. Thus, epigenetic modifications associated with methylation of the DNA and histones are accompanied by changes in gene expression in the presence of signals from the external environment. In particular, this is manifested in models of various muscles loadings.
An important step in understanding of the role of the DNA methylation in the gene expression in the skeletal muscles was the study performed on different types of human muscle fibers [7]. Various DNA methylation sites have been identified in slow and fast types of muscle fibers. The expression of a number of structural and metabolic genes has been shown. Difference in methylation levels of DNA affects metabolism in specialized types of muscle fibers in humans. Thus, hypermethylation of genes involved in anaerobic metabolism was revealed in slow muscle fibers. These are the following genes: rapid calcium pump (SERCA1), phosphofructokinase (PEKM), aldolase A (ALDOA) and uncoupling protein 3 (UCP3). On the other hand, hypermethylation of genes that participate in aerobic metabolism, such as the slow calcium pump (SERCA2) and lactate dehydrogenase B (LDH-H) was revealed in fast muscle fibers. The method of determination of methylation sites in specialized muscle fibers proposed by the authors provides new possibilities for discovering molecular differences in heavy myosin chains in skeletal muscle fibers of types I and IIa.
Exercising causes a change in the methylation of promoters of metabolic and structural genes in the skeletal muscles. Thus, methylation of the promoters of the genes PPARGC1A, TFAM, MEF2A, CS and PDK4 [6] decreases after a single physical load in human muscles. Numerous regulating pathways participate in the methylation process of the genes promoters in the skeletal muscles. These pathways regulate the following processes: release of Ca2+, enhancing oxidative phosphorylation and increasing the ATP production, changing the ratio of AMP:ATP, activation of 5-AMP proteinkinase (AMPK) and increase in reactive oxygen species, change in the S-adenosylmethionine level, the main supplier of methyl groups, as well as the possible enhancement of demethylation reactions involving TET/BER enzymes.
Six months training program led to a change in the DNA methylation profile in the skeletal muscles of the subjects. Most of the studied genes were at a different stage of hypomethylation after the training [8]. Therefore, it convincingly shows that single and systematic physical activity is accompanied by changes in DNA methylation in the skeletal muscles which is the most significant epigenetic factor. Action-oriented games at an early age as well as active physical training and sports of schoolchildren and students are able to maintain human skeletal muscles in the stable expression regime of the genes responsible for fat oxidation. Otherwise, in people with a sedentary lifestyle, the metabolic-favorable expression of genes can be stubbornly suppressed by epigenetic mechanisms increasing the risk of obesity. From infancy, parents have a risk “to program” the future obesity of their children, if parents influence physical activity of children and the amount / quality of food they consume. Moreover, humans can transmit a pathological epigenetic program to their descendants while having a normal DNA structure.
Two types of enzymes control the reversibility of the acetylation of amino acid residues of lysine in the histone molecule in the post-translational modification of proteins: histone acetylase (HTA) and histone deacetylase (HDAC). These enzymes catalyze the transfer of the acetyl group to the amino acid lysine in the N-terminal region of the histone molecule H2A, H2B, H3 and H4. At present, 18 HDAC enzymes have been identified and divided into 4 classes. Enzymes included in class IIa are involved in the regulation of skeletal muscle metabolism. These enzymes differ a lot from other classes of histone deacetylases. Primarily because they are tissue specific and involved in the regulation of the skeletal and cardiac muscle metabolism. Enzymes of class IIa participate in reactions with specific transcription factors, and their signal-dependent nuclear-cytoplasmic movement has an effect on the regulation of genes in skeletal muscles [1].
Quiescent HDAC enzymes of class IIa are located in the nucleus in a dephosphorylated state and interact with transcription factors and regulatory proteins as gene activity regulators. HDAC phosphorylation takes place when information signals are transferred into the nucleus. This leads to the disintegration of protein complexes with transcription factors followed by enzymes release into cytoplasm.
Regulation of metabolism in skeletal muscles when performing physical activity of different intensity and duration occurs with the participation of HDAC enzymes of class IIa. The role of these enzymes was studied on different models both with a single and systematic physical action. Phosphorylation of HDAC and transport of the enzyme from the nucleus promote the enhancement of the transcriptional response during physical exercise. Consequently, the group of HDAC enzymes can be considered as the key regulators of metabolism in the skeletal muscles, controlling the processes of metabolic adaptation to external influences including physical activity [1].
In recent years, the facts appeared showing that the connection between epigenetics and metabolism is two-way involving metabolites from the common metabolic pathways used to modify proteins and the DNA. A key factor in the regulation of the skeletal muscle metabolism is the transcriptional control of the metabolic enzymes expression. This aspect can be regarded as the main one for epigenetic regulation. As a confirming example, presence of acetyl-CoA in the cytoplasm and nucleus of muscle cells can be cited, since it serves as a substrate in the protein acetylation reaction. With a presence of enzymes acetyltransferases and deacetylases in the muscle cell the process is regulated by a high level of acetyl-CoA. Low concentration of this substrate makes the process ineffective. Thus, the availability of acetyl-CoA for histone acetylation largely determines the intensity of gene expression and the metabolism level in skeletal muscles, both at rest and in functional activity. The increase in enzyme ATP citrate lipase (ACL) activity in muscles increases the metabolism in the mitochondria. Production of ATP increases at the same time. Therefore, increase of the availability of acetyl-CoA leads to increase in skeletal muscle metabolism.
In recently published article by Australian scientists an issue of inheritance of skeletal muscle metabolic phenotypes is discussed [9]. Discussion is focused on changes in the skeletal muscle metabolism caused by post-translational modifications of histones in acetylation / deacetylation and DNA methylation reactions in models on cell cultures, animal experiments and human physical activity. Analysis of the obtained data made it possible to establish that the key factor for the regulation of the skeletal muscle metabolism is the transcriptional control of the metabolic genes expression. DNA methylation and histone modification in acetylation and methylation reactions, as well as the participation of microRNAs, play an important role in the epigenetic regulation of gene expression and skeletal muscle metabolism in various functional states of a human organism.
The classical triad of RNA components of protein synthesis (informative, transport and ribosomal) has gradually expanded, and new noncoding RNA proteins with different functions were detected. A decisive breakthrough occurred in the early 2000s, when a class of small RNAs, microRNAs, containing ~ 22 nucleotides, was discovered. This group of noncoding protein RNA takes part in the regulation of many cellular processes in different representatives of flora and fauna. MicroRNAs implement posttranscriptional silencing of genes – RNA interference. They are able to regulate the intensity of transcription processes, RNA processing and translation by complementary interaction with DNA or mRNA in different human organs and tissues. According to the bioinformative estimation of microRNA targets, up to 60% of all human genes are controlled by microRNA. Each mRNA has binding sites to many mRNAs, and one mRNA represents an optional target for several mRNAs. Thus, microRNAs and mRNAs form a complex network of regulatory interactions that participate in the epigenetic modification of gene expression [2].
In the skeletal muscles, tissue-specific microRNAs are miR-1, miR-133a, miR-133b, miR-206, which have been termed myomiRNA. These four miRNAs belong to the miR-1 family, which can be divided into two groups, miR-1/206 and miR-133a/b, based on differences in specific sequences in the molecule structure. It should be noted that microRNA regulates the basic functions of the skeletal muscles, such as myogenesis, energy metabolism, protein synthesis, hypertrophy and atrophy. The myomiRNA family is gradually expanding and currently includes 7 microRNAs: 1, 133, 206, 208a, 208b, 486, 499. Studies performed on animals and humans of different ages using different types of exercise showed the involvement of epigenetic modification in structural and metabolic genes expression in the skeletal muscles. The use of single and systematic physical loads of different energy directions allowed not only to detect new myomiRNA, but also to determine the intensity of genes expression involved in the skeletal muscle metabolism.
During systematic aerobic exercises, the level of microRNA expression in the skeletal muscles increases. It is accompanied by increase of mitochondria biogenesis and enzymes concentration that provides aerobic metabolic reactions and causes dilatation of the capillary network. Each microRNA has many targets and this makes it difficult to determine the effect of individual molecules under different metabolism conditions of tissues and human organs. The reaction of microRNAs in the human body to different external influences differs significantly. It is confirmed by the results obtained after different types of exercise and using different nutrients. It can be assumed that as the range of studies will be expanded, microRNAs will be used as epigenetic markers of different genotypes. In particular, they will be used for revealing skeletal muscle damage and evaluating adequacy of a body's response in terms of physical activity to a human functional state [2].
Further research of development, formation and improvement of human physical qualities will be related to molecular determinants which will include genetic and epigenetic factors. Combining and analyzing the data of genomics, epigenomics and transcriptomics with the help of bioinformatic tools will enable us to expand understanding of the molecular mechanisms that regulate metabolism in human skeletal muscles during physical exertions of different intensity and duration. A wide range of skeletal muscle metabolism during exercises of different intensity and duration creates opportunities for improvement of functional activity as well as further and deeper study of the molecular mechanisms of epigenetic regulation.
References
- Astratenkova I.V., Rogozkin V.A. Rol atsetilirovaniya /deatsetilirovaniya gistonov i transkriptsionnykh faktorov v regulyatsii metabolizma skeletnykh myshtsakh [The role of acetylation/deacetylation of histones and transcription factors in the regulation of skeletal muscle metabolism]. Rossiyskiy fiziologicheskiy zhurnal im. I.M. Sechenova, 2017, vol. 103, no. 6, pp. 593-605.
- Astratenkova I.V., Rogozkin V.A. Uchastie mikroRNK v regulyatsii metabolizma skeletnykh myshts [The role of microRNA in the regulation of skeletal muscle metabolism]. Rossiyskiy fiziologicheskiy zhurnal im. I.M. Sechenova, 2015, vol. 101, no. 7, pp. 753-772.
- Akhmetov I.I. Molekulyarnaya genetika sporta [Sports molecular genetics]. Moscow: Sovetskiy sport publ., 2009, 268 p.
- Rogozkin V.A., Nazarov I.B., Kazakov V.I. Geneticheskie markery fizicheskoy rabotosposobnosti cheloveka [Genetic markers of individual physical working capacity]. Teoriya i praktika fiz. kultury, 2000, no. 12, pp. 33-36.
- Rogozkin V.A. Rasshifrovka genoma cheloveka i sport [Decoding of human genome and sport]. Teoriya i praktika fiz. kultury, 2001, no. 6, pp. 60-63.
- Barres R., Yan J., Edan B., Treebak J.T., Rasmunssen M., Fritz T., Caidahl K., Krook A., O’Gorman DJ., Zierath J.R. Acute exercise remodels promoter methylation in human skeletal muscule. Cell Metab., 2012, 15, no. 3, pp. 405-411.
- Begue G., Rane U., Jemiolo B., Trappe S. DNA mutilation assessment from human slow-and fast-twitch skeletal muscule fibers. J Apply Physiol (1985), 2017. 122, no. 4, pp.952-967
- Hoffman N.J. Omics and Exercise: Global Approches for Mapping Exercise Biological Networks. Cold Spring Harbor Persrect Med., 2017. 7.a 029884
- Howletd K.F., McGee S.L. Epigenetic regulation of skeletal muscle metabolism. Clinical Science, 2016. 130, pp. 1051-1063.
- Pitsiladis Y.P., Tanaka M., Eynon N., Bouchard C., North K.N., Williams A.G., Collins M., Moran C., Britton S.L., Fuku N., Ashely E.A., Klissouras V., Lusia A., Ahmetov I.I., de Gens E., Alsayrafo M. Athlome Project Consortium: a concerted effort to discover genomic and other “omic” markers of athletic performance. Physiol. Genomics, 2016, vol.48, pp.183-190.
- Sharples A.P., Stewart C.E., Seaborne R.A. Does skeletal muscle have an epi-memory? The role of epigenetics in nutritional programming< metabolic disease, aging and exercise. Aging Cell, 2016, 15, pp. 603-616.
Corresponding author: ndgolberg@gmail.com
Abstract
A complex set of molecular mechanisms, including genetic and epigenetic factors, is involved in the regulation of skeletal muscle metabolism. Cascades of reactions associated with the proteins posttranslational modification are a universal mechanism for controlling intracellular metabolism, which provides development, differentiation, transformation, hypertrophy and atrophy of muscle cells. Epigenetic regulation of gene expression in human skeletal muscles is carried out at three levels: DNA methylation, histone modification and mRNA–microRNA interaction. The article considers the possible mechanisms of epigenetic regulation of intracellular metabolism of skeletal muscles during single and systematic physical exercises. Main focus is given to the epigenetic regulation of structural and metabolic genes expression in skeletal muscles at different functional states of a human organism.



 Журнал "THEORY AND PRACTICE
Журнал "THEORY AND PRACTICE