Genotype and metabolic profile associations in athletic adaptation to physical loads
ˑ:
D.A. Alaverdyan1
PhD, Associate Professor S.Sh. Namozova2
Dr.Med., Professor S.G. Shcherbak1, 2
S.P. Urazov2
PhD O.S. Glotov1, 3
1City Hospital № 40, Sestroretsk
2St. Petersburg State University, St. Petersburg
3D. O. Ott Research Institute of Obstetrics and Gynecology, St. Petersburg
Keywords: sport genetics, metabolism related genes, polymorphism, adaptation, treadmill test.
Introduction. Studies of energy metabolism specifics in an athlete’s body under heavy varied-intensity physical workloads made it possible to find the genetic determinants associated with the training process. Trainings cause metabolic changes in the bodily systems, for instance, the muscle capacity for lipid and carbohydrates oxidation increases. One-time physical load causes changes in the expression of hundreds of genes, which reverts with time (seconds, minutes, hours). Therefore, it can be argued that long-term adaptation to physical loads is formed owing to a series of short-term metabolic changes caused by repeated trainings, which lead to global changes in the gene expression regulatory system. Adaptation to physical loads is due to regular trainings coupled with the level of expression of many genes, which, in turn, varies depending on nucleotide substitutions in the coding regions of the genes. The mechanisms of formation of adaptation to physical loads were studied based on the selected genetic markers that could have a direct or indirect effect on metabolism during physical activity [1, 4].
Objective of the study was to identify the genotype and metabolic profile associations in athletic adaptation to physical loads.
Methods and structure of the study. Sampled for the study were 71 athletes specialized in 11 sports, including 69 student-athletes of mass categories and 2 masters of sport. The mean age of the sample at the time of the experiment was 20 years. This group cannot be attributed to that of elite sportsmanship, however, the initial blood pressure (BP) values are slightly different from the data obtained in the individuals of the same age who regularly attend physical education classes but do not do any particular sport, which indicates a certain level of adaptation to physical loads or physical fitness level. The subjects were warned about the experimental conditions and gave a voluntary written consent to participate in it. The experiment was approved by the Ethics Committee of St. Petersburg State University.
The study was carried out in a clinical setting on the basis of the State Budgetary Healthcare Institution "The City Hospital No 40 of St. Petersburg" and involved the following clinical and instrumental methods of examination aimed to assess the cardiovascular system functionality rates: electrocardiography using the MAK 2000 device (GE, USA), daily Holter ECG monitoring using the wrist-worn activity meters Cardiotechnics-4 (Incart, Russia), echocardiography using the Vivid E9 XDclear 4D device (GE, USA), a treadmill test according to the BRUCE Protocol while measuring the blood pressure level and heart rate.
The sample of athletes were grouped in three different ways:
1. Males (N=47) and females (N=24).
2. With the high (14.1-23.9 MET) and low (7-14 MET) values of metabolic equivalent (MET) based on the data obtained in BRUCE Treadmill Test [6].
3. In accordance with the classification of sports by the type of impact of training loads (see Table 1).
Table 1. Classification of sports by the type of impact of training loads
|
№ |
Classification of sports by the type of impact of training loads |
Number of sports |
Number of people |
Kind of sport |
|
1 |
Cyclic sports |
1 |
5 |
Rowing |
|
2 |
Sports games |
5 |
38 |
Football, volleyball, hockey, badminton, rugby |
|
3 |
Complex coordination sports |
2 |
13 |
Dance sport, cheerleading |
|
4 |
Complex technical sports and all-round sports* |
3 |
15 |
Mountaineering, sport orienteering, triathlon |
* – neuropsychic strain is a limiting factor in the development of physical working capacity.
The DNA extraction from the peripheral blood leukocytes was carried out by the standard chloroform extraction method using 5M NaCl. The functionally significant polymorphic sites of the following genes were studied by means of the PCR-RFLP analysis: ACE - a gene that regulates blood pressure, PPARA, PPARD, PPARG – genes involved in the exchange of cholesterol, carbohydrates and oxidation of fatty acids, PPARGC1A - a gene encoding a coactivator of transcriptional factors and estrogen and mineralocorticoid receptors, AMPD1 - a gene responsible for the energy metabolism in the skeletal muscles during muscle activity, ACTN3 - a gene that determines the type of the muscle fibers, as well as the genes of dopamine and serotonin receptors: DRD2A, HTR2A.
Results and discussion. We compared the frequency of alleles (genotypes) in the sample of athletes (N=71) with those in the population of St. Petersburg [2, 3, 5] and Europe (N=503, according to www.ensembl.org) using a chi-square test.
The intragroup comparison of the allele frequencies and matching of the obtained data with the population sampling revealed no statistically significant differences (p>0.4). Such a result may be due to the specificity of the sample: perhaps, the examined athletes were not skilled enough to differ significantly from the population. The analysis of the genotype frequencies in the groups of athletes with different fitness levels did not reveal any statistically significant differences (p>0.1). This can be explained by the small sample size (N=71) or the fact that the polymorphisms of the studied genes that we selected are not associated with MET. No significant age-specific differences were found between the groups either (p>0.1).
The analysis of the clinical and laboratory data revealed increased levels of creatine kinase (319.7 mmol/L) and homocysteine (14.6 mmol/L) in the male athletes as opposed to the normal values in healthy males (<190 mmol/L and 5.5-12.2 mmol/L, respectively), which may be due to regular physical activity.
We analyzed the associations of the clinical and laboratory data with the studied gene polymorphism. The athletes with different polymorphic variants of PPARGC1A gene were found to have statistically significant differences in blood cholesterol (p=0.004), triglycerides (p=0.042) and high density lipoproteins (HDL) (p=0.012), the level of which in PPARGC1A A/A (Ser/Ser) genotype carriers was significantly higher than in G/G (Gly/Gly) and G/A (Ser/Gly) genotype carriers. It is known that the expression of PPARGC1A gene significantly increases during physical activity [4]. The studied gene polymorphism is associated with the changes in the level of oxidation of fatty acids, functions of the vascular and respiratory systems, as well as insulin sensitivity of the liver cells and skeletal muscles. PPARGC1A G/G and G/A genotype carriers were found to have more intense oxidation of fatty acids in the liver, myocardium, and skeletal muscle as compared to A/A genotype carriers [8]. The findings confirm that PPARGC1A A/A genotype carriers are characterized by decreased metabolism of cholesterol, triglycerides and HDL.
The study also found that in athletes with ACE D/D genotype, the level of resting diastolic pressure is higher as opposed to I/D and I/I genotype carriers (p=0.017, see Figure 1).
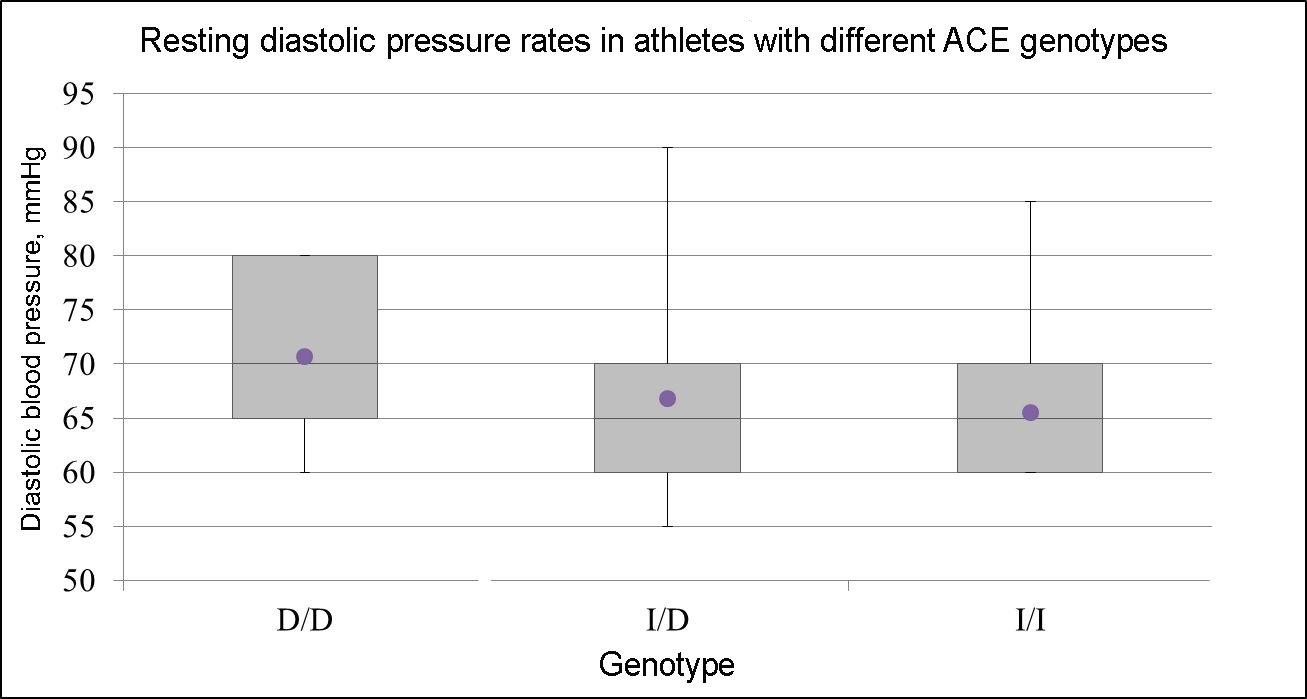
Figure 1. Resting diastolic pressure (mmHg) rates in athletes with different ACE genotypes.
D/D genotype (rs4340) is associated with higher blood pressure (p-value=0.017). D/D (N=22), I/D (N=30), I/I (N=19)
The ACE gene polymorphism has been well studied in the context of blood pressure regulation. We have confirmed the functional association of ACE gene with blood pressure rates: D/D genotype causes higher blood pressure. In addition, it should be noted that D/D genotype is associated with an increased risk of cardiovascular disease [7], which can be especially dangerous during sports activities.
Conclusion. The detected functional relationships between the metabolism controlling genes and blood biochemistry values, that confirm and supplement the literature data, indicate that the studied polymorphism does play a key role in the formation of adaptation to physical loads and affect the level of gene expression. Since the experiment was conducted on a relatively small sample, its data need to be verified on a larger volume of material to detect new significant functional relationships between the genotype and physiological test rates.
References
- Alaverdyan D.A., O.P. Mamaeva, S.Sh. Namozova et al. Analiz pokazateley metabolicheskogo profilya i polimorfizma genov ACE, PPARA, PPARD, PPARG, PPARGC1A, AMPD1, ACTN3, DRD2A, HTR2A pri adaptatsii k razlichnym vidam fizicheskoy nagruzki [Analysis of metabolic profile and polymorphism of ACE, PPARA, PPARD, PPARG, PPARGC1A, AMPD1, ACTN3, DRD2A, HTR2A genes when adapting to various types of physical activity]. Molekulyarno-biologicheskie tekhnologii v meditsinskoy praktike [Molecular Biological Technologies in Medical Practice]. A.B. Maslennikov [ed.]. no. 27. Novosibirsk: Akademizdat, 2018. pp. 84-96.
- Glotov A.S., Glotov O.S., Moskalenko M.V. et al. Analiz polimorfizma genov renin-angiotenzinovoy sistemy v populyatsii Severo-Zapadnogo regiona Rossii, u atletov i u dolgozhiteley [Analysis of polymorphism of genes of renin-angiotensin system in population of North-Western region of Russia, in athletes and long-livers]. Ekologicheskaya genetika. 2004.no. 4. pp. 40-43.
- Glotov O.S., Glotov A.S., Vakharlovskiy V.G. et al. Izuchenie polimorfizma genov ACE, AGT, AGTR1, NOS3, MTHFR, GPIIIa, PAI-1 v treh vozrastnyih gruppah Severo-Zapadnogo regiona Rossii [Study of ACE, AGT, AGTR1, NOS3, MTHFR, GPIIIa, PAI-1 gene polymorphisms in three age groups of North-West region of Russia]. Donozologiya i zdorovy obraz zhizni. 2010. no. 2 (7). pp. 7-11.
- Glotov O.S., Glotov A.S., Pakin V.S. et al. Monitoring zdorovya cheloveka – vozmozhnosti sovremennoy genetiki [Monitoring of human health - modern genetics potential]. Vestnik Sankt-Peterburgskogo universiteta. 2013. ser. 3. no.. 2. pp. 95-107.
- Potulova S.V., Glotov O.S., Baranov V.S. Analiz polimorfizma genov IGF1 i PGC1, uchastvuyuschikh v energeticheskom obmene, v dvukh vozrastnykh gruppakh naseleniya Sankt-Peterburga [Analysis of polymorphism of IGF1 and PGC1 genes involved in energy metabolism in two age groups of St. Petersburg population]. Ekologicheskaya genetika. v. VII, no. 1. 2009. pp.. 12-18.
- Tavrovskaya T.V. Veloergometriya [Bicycle ergometry]. Doctor's guide. St. Petersburg, 2007. P. 134.
- Puthucheary Z., Skipworth J.R., Montgomery H.E. The ACE gene and human performance: 12 years on. Sports Med. 2011. 41 (6): 433-48.
- Rankinen T., Bray M.S., Hagberg J.M., Perusse L., Roth S.M., Wolfarth B., Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2005 update. Med Sci Sports Exerc. 2006. V.38 (11). pp.1863-1888.
Corresponding author: Ditanka17@gmail.com
Abstract
Studies of energy metabolism specifics in athlete’s body under heavy varied-intensity physical workloads made it possible to find the genetic determinants associated with the training process. Different genes are known to be of influence on the key functions of energy metabolism. Frequency of alleles favorable for some physical quality building (e.g. endurance) is higher in athletes versus untrained people and may reach maximums in the sport leaders. Sampled for our study of the genetic and biochemical factors of influence on the athletic adaptation to physical workloads were 71 sporting students aged 20 on average and specialized in 11 sports. We used the PCR-PDRF analysis to identify genotypes for 9 genes including ACE (rs4340), PPARA (rs4253778), PPARD (rs2016520), PPARG (rs1801282), PPARGC1A (rs8192678), AMPD1 (rs17602729), ACTN3 (rs1815739), DRD2A (rs1800497) and HTR2A (rs6313). We also tested the cardiovascular system functionality and blood biochemistry of the sample. The study found no meaningful intergroup differences in the trained versus untrained groups. We confirmed and supplemented the functional associations of PPARGC1A and ACE genes with the physiological and biochemical test rates to discuss the gene polymorphism triggered by physical trainings.


 Журнал "THEORY AND PRACTICE
Журнал "THEORY AND PRACTICE